Howard University Department of Biology Assistant Professor Michael C. Campbell and an international team of scientists led by members of Professor Sarah Tishkoff’s lab at the University of Pennsylvania have identified new variants in or near the SLC24A5, MFSD12, DDB1, TMEM138, OCA2 and HERC2 genes that are responsible for skin color in modern humans.
Unlike prior skin-color-research studies, which have focused on European subjects, the present study analyzed genetic data from a large set of 1,570 people from populations in very remote regions in Ethiopia, Tanzania and Botswana located in eastern and southern Africa, making it the first of its kind. The individuals included in this study practice different modes of subsistence and possess a broad spectrum of skin pigmentation present in Africa.
Among the partnering local academic institutions were the University of Botswana, University of Addis Ababa in Ethiopia, and Muhimbili University of Health and Allied Sciences in Tanzania. As part of the process of data collection, public meetings with local authorities, community members, translators and the researchers from the University of Pennsylvania were held to ensure that participants were fully and properly informed about the nature of the study.
“In Botswana we collected genetic and phenotype data from San hunter-gatherers who have lighter pigmentation compared to their Bantu-speaking neighbors,” said Campbell. “In Tanzania and Ethiopia, genetic and phenotype data were collected from Nilo-Saharan and Bantu-speaking populations that have darker pigmentation.”
The study also collected a range of phenotypic information, including skin melanin index which is a measure of melanin content, from over 2,000 ethnically and genetically diverse subjects. According to Campbell, by analyzing both genetic and phenotypic information, researchers identified novel alleles—or variants—that explain approximately 30 percent of the skin pigmentation differences in African populations, which is considered high for a human variable trait.
“Because alleles in or near the genes identified in our study account for a large proportion of the differences in skin pigmentation, we argue that there are fewer genes of strong effect that influence this trait, whereas for a trait like human height, for example, there are likely a larger number of alleles that contribute to observable differences,” said Campbell.
Campbell also noted a particular strength of the study is the functional follow-up across animal models, such as the zebrafish and mouse, which provided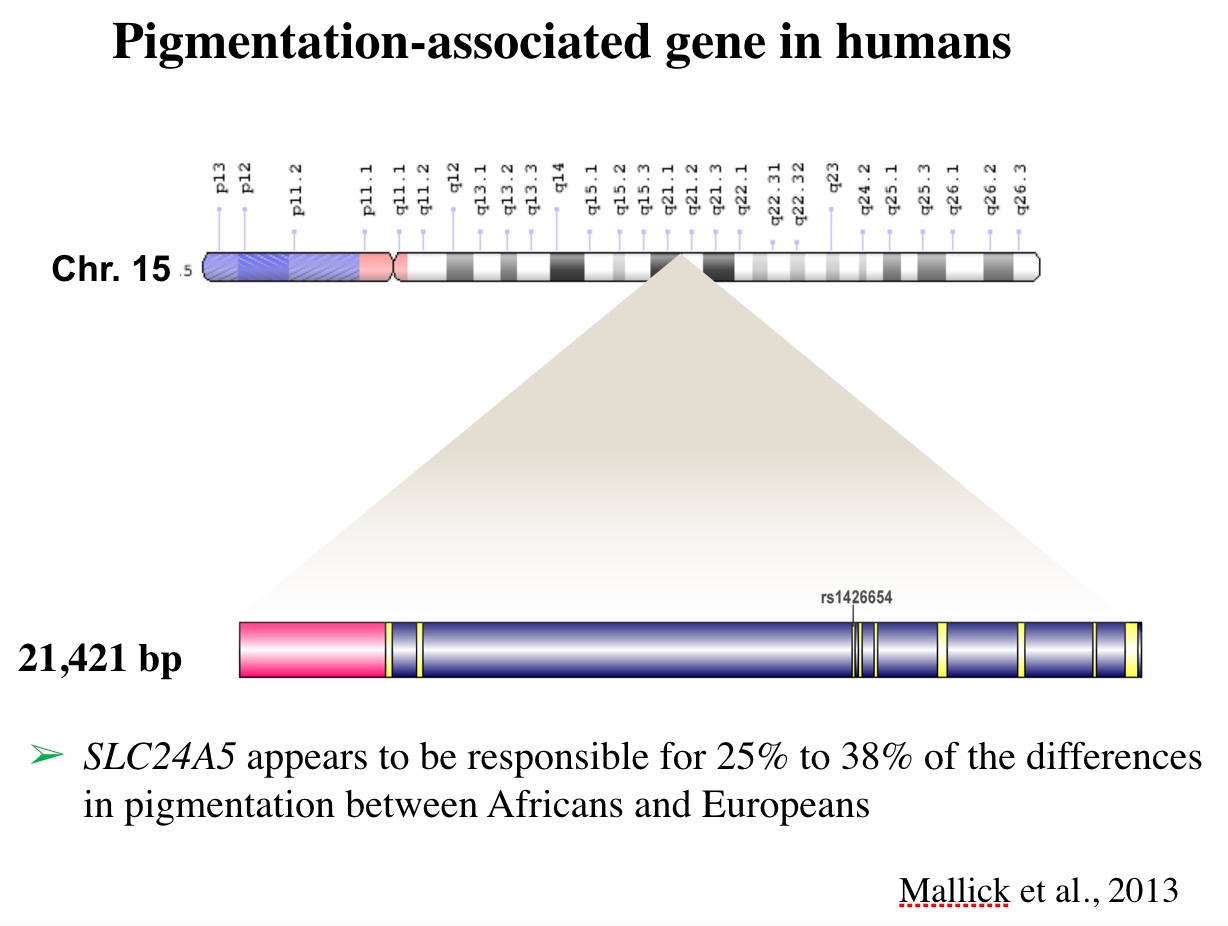 very strong evidence that the alleles, identified through the researchers’ genome-wide association study (GWAS) approach, in fact played a role in skin color determination and were not false positive association hits. Zebrafish are a particularly relevant organism since they were instrumental in the identification of the human SLC24A5 gene, which was found to contribute to light skin in populations of European and some south Asian populations in previous studies.
very strong evidence that the alleles, identified through the researchers’ genome-wide association study (GWAS) approach, in fact played a role in skin color determination and were not false positive association hits. Zebrafish are a particularly relevant organism since they were instrumental in the identification of the human SLC24A5 gene, which was found to contribute to light skin in populations of European and some south Asian populations in previous studies.
“One of the other striking findings of our research is that many of the alleles associated with darker pigmentation are derived, as opposed to ancestral as previously believed,” said Campbell. “Furthermore, alleles for lighter and darker skin pigmentation are both present in Africa—alleles associated with lighter skin pigmentation are found at higher frequency in the San hunter-gatherers from southern Africa, while allele associated with darker skin pigmentation are found at higher frequency in Nilo-Saharan and other sub-Saharan Africans except the San. Most of these alleles for light and dark pigmentation are also very old (up to hundreds of thousands of years old in Africa), predating the origin of modern humans. One implication of this finding is that it indicates lighter pigmentation did not evolve recently outside of Africa but was already present within Africa for a long period of time.”
“In addition, the same genetic variants (or alleles) correlated with darker skin pigmentation in Africans are the same as the variants correlated with dark skin pigmentation in South Asian and Australo-Melanesian populations,” said Campbell. These finding indicate that the alleles contributing to dark pigmentation in South Asians and Australo-Melanesians were inherited from Africans, and did not evolve independently. One of the strongest implications of this finding is that it challenges notions of race which has historically been defined by skin color and establishes clear genetic relationships between different human populations.
“Sociologically and historically, value has been placed on skin pigmentation differences and these differences have been at the root of injustices committed against individuals over an extended period of time,” said Campbell. “However, our study highlights the fact that this biological trait, like a host of others in our species—[such as] lactase persistence or bitter taste perception—is a normal variable trait that arose in response to environmental factors during human evolutionary history.”
Most notably, both light and dark skin pigmentations have conferred benefits to humans. In particular, darker skin is believed to help protect against the negative impacts of ultraviolet light exposure, while lighter skin helps to promote synthesis of vitamin D, which requires sunlight to penetrate the skin, in regions of the world with low ultraviolet light exposure.
For Campbell, the only researcher from an HBCU represented amongst the 47 who contributed to the study, this research is of particular interest, because of its insights into understanding the genetic basis and evolutionary history of skin pigmentation—particularly darker pigmentation—and its ability to reveal the history of people who are highly underrepresented in human genomic and evolutionary studies.
According to Campbell, given that existing African samples included in the study, while comprehensive, are not exhaustive, one future direction of this research could involve sampling individuals in other regions in Africa, such as West and West Central Africa, for a more complete geographic representation of diverse Africans in the overall dataset.
“One other possible future study could be to examine genomic variation in African Americans to better understand the extent to which the novel alleles identified in our study contribute to skin pigmentation in this recently admixed population,” said Campbell. “Additionally, it would be very interesting to test for an association between the genomic regions identified in our study and skin diseases in Africans, such as vitiligo, or congenital conditions, such as albinism.”
“Indeed, [this] study provides a great starting point for investigating whether or not individuals with certain genetic variants are more or less susceptible to DNA damage caused by ultraviolet radiation or respond to cellular stress differently.”
For the full findings of the study in the journal Science, please visit http://science.sciencemag.org/content/358/6365/eaan8433.
Watch Campbell's video interview with TheAfricanDream LLC's Oral Ofori at https://www.youtube.com/watch?v=Pz8jliHipqU&spfreload=5.
--------
*Head-shot by: Howard University
**Gene structure of SLC24A5 by: Basu Mallick C, Iliescu FM, Möls M, Hill S, Tamang R, Chaubey G, et al. (2013) The Light Skin Allele of SLC24A5 in South Asians and Europeans Shares Identity by Descent. PLoS Genet9(11): e1003912
***Collage by: Alessia Ranciaro and Simon Thompson